Sliced traversals
Writing Monte Carlo simulation can be a fiddly task. For one, it’s sometimes hard to follow parametrizations mentioned (not explained) in existing literature. Sometimes, your simulation is just not saturating the right statistic.
Another thing is getting your code to run quickly to jump across orders of magnitudes of domain sweeps. Depending on your algorithmic formulation you’re allowed to do sequential trials. Sometimes you may divide your domain into subdomains which exchange halos and thus you can parallelize nicely. Some people spend a long time trying to come up with with patterns that let your architecture perform sweeps in an embarrassingly parallel setting.
The Hamiltonian I’m currently looking at is defined on a cubic lattice. Every term is local in that it only depends on its nearest neighbors. Translating this into a Monte Carlo formulation already poses the problem of having rather large radii for each trial move you propose: Threads running on the same grid will need to be at least 4 steps apart (in each dimension) or else you’re introducing a race condition.
Here’s a tedious but I’d say somewhat safe way to go about parallelizing across the grid: Assume your grid has dimensions L x L x L: Fix indices i + t * 4 for each thread t. Then, each thread can comfortably traverse its own 2d slice with (j, k) in [0, L) x [0, L) of the grid in parallel to all other threads t’. Unfortunately, this implies you would have to do this 4 times (modulo L). Hence you would need to introduce about 4 barriers for each sweep you take. You want to minimize waits and thread spawns, so running the parallelization in an already parallel region with well-defined parameters should do the trick.
As an illustration, consider the following pythonic grid:
L = 12
N = L * L * L
x = np.arange(0, L)
y = np.arange(0, L)
xx, yy = np.meshgrid(x, y)
plt.hlines(y, 0, L, color='k', alpha=.2)
plt.vlines(x, 0, L, color='k', alpha=.2)
plt.scatter(xx, yy, color='w', marker='s', s=50)
colors = ["red", "green", "blue", "pink"]
for s in range(4):
for t in range(4):
for i in range(s, L, 4):
for j in range(t, L, 4):
plt.scatter(xx[i, j], yy[i, j], color=colors[t])
We create a grid and color the slices which we can process in parallel with the same colors:
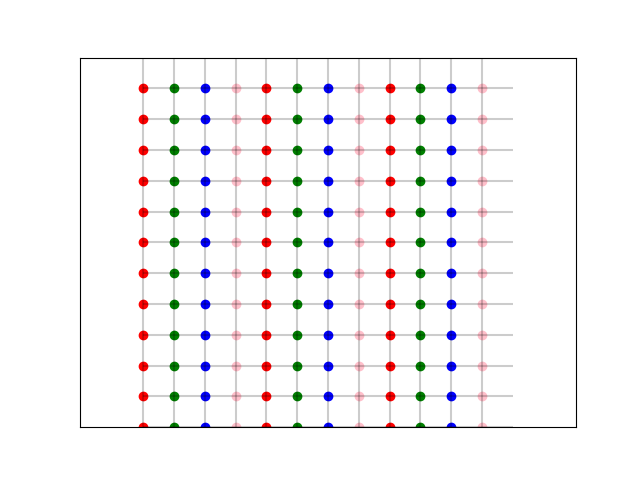
Accessing the nearest neighbors will be done like so.
def nn(i, j, L):
l, r = (i, (j - 1) % L), (i, (j + 1) % L)
u, d = ((i - 1) % L, j), ((i + 1) % L, j)
return l, r, u, d
Maybe I’ll expand this with a more illustrative Jupyter notebook.